Note
Go to the end to download the full example code or to run this example in your browser via Binder
Getting the data#
In this section, we will dicuss how to get functional data to
use in scikit-fda. We will briefly describe the
FDataGrid class, which is the type that
scikit-fda uses for storing and working with functional data in discretized
form. We will discuss also how to import functional data from several sources
and show how to fetch and load existing datasets popular in the FDA
literature.
# Author: Carlos Ramos Carreño
# License: MIT
#
# sphinx_gallery_thumbnail_number = 6
The FDataGrid class#
In order to use scikit-fda, first we need functional data to analyze.
A common case is to have each functional observation measured at the same
points.
This kind of functional data is easily representable in scikit-fda using
the FDataGrid class.
The FDataGrid has two important
attributes: data_matrix and grid_points.
The attribute grid_points is a tuple with the same length as the
number of domain dimensions (that is, one for curves, two for surfaces…).
Each of its elements is a 1D numpy ndarray containing the
grid points for that particular dimension,
where \(M_i\) is the number of measurement points for each “argument” or domain coordinate of the function \(i\) and \(p\) is the domain dimension.
The attribute data_matrix is a
numpy ndarray containing the measured values of the
functions in the grid spanned by the grid points. For functions
\(\{x_n: \mathbb{R}^p \to \mathbb{R}^q\}_{n=1}^N\) this is a tensor
with dimensions \(N \times M_1 \times \ldots \times M_p \times q\).
In order to create a FDataGrid, these
attributes may be provided. The attributes are converted to
ndarray when necessary.
Note
The grid points can be omitted,
and in that case their number is inferred from the dimensions of
data_matrix and they are automatically assigned as equispaced points
in the unitary cube in the domain set.
In the common case of functions with domain dimension of 1, the list of
grid points can be passed directly as grid_points.
If the codomain dimension is 1, the last dimension of data_matrix
can be dropped.
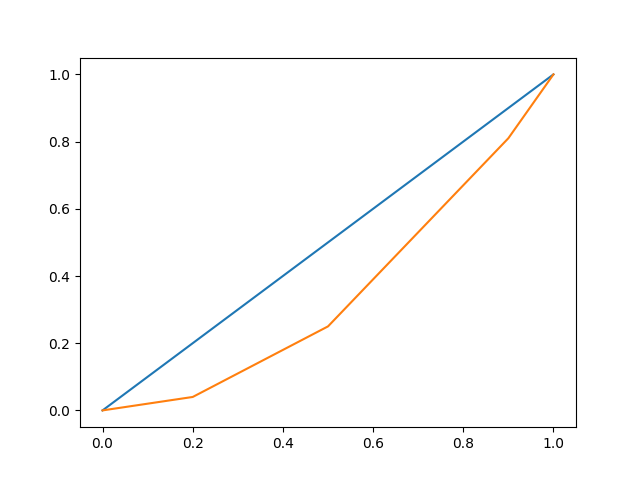
The following example shows the creation of a
FDataGrid with two functions (curves)
\(\{x_i: \mathbb{R} \to \mathbb{R}\}, i=1,2\) measured at the same
(non-equispaced) points.
import skfda
import matplotlib.pyplot as plt
grid_points = [0, 0.2, 0.5, 0.9, 1] # Grid points of the curves
data_matrix = [
[0, 0.2, 0.5, 0.9, 1], # First observation
[0, 0.04, 0.25, 0.81, 1], # Second observation
]
fd = skfda.FDataGrid(
data_matrix=data_matrix,
grid_points=grid_points,
)
fd.plot()
plt.show()
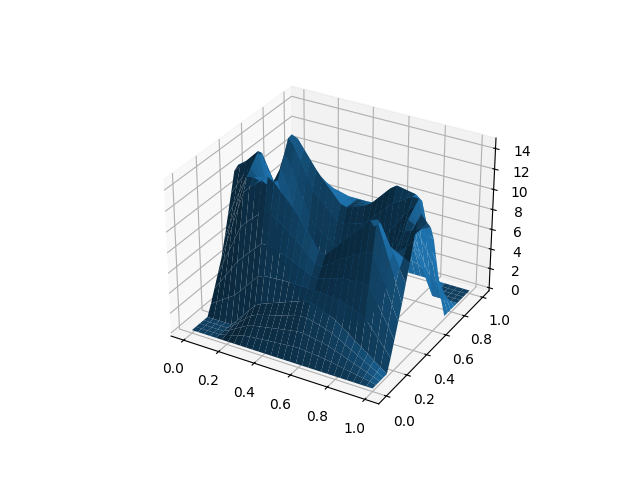
Advanced example#
In order to better understand the FDataGrid structure, you can consider the
following example, in which a FDataGrid
object is created, containing just one function (vector-valued surface)
\(x: \mathbb{R}^2 \to \mathbb{R}^4\).
grid_points_surface = [
[0.2, 0.5, 0.7], # Measurement points in first domain dimension
[0, 1.5], # Measurement points in second domain dimension
]
data_matrix_surface = [
# First observation
[
# 0.2
[
# Value at (0.2, 0)
[1, 2, 3, 4.1],
# Value at (0.2, 1.5)
[0, 1, -1.3, 2],
],
# 0.5
[
# Value at (0.5, 0)
[-2, 0, 5.5, 7],
# Value at (0.5, 1.5)
[2, 1.1, -1, -2],
],
# 0.7
[
# Value at (0.7, 0)
[0, 0, 1.1, 1],
# Value at (0.7, 1.5)
[-3, 5, -0.5, -2],
],
],
# This example has only one observation. Next observations would be
# added here.
]
fd = skfda.FDataGrid(
data_matrix=data_matrix_surface,
grid_points=grid_points_surface,
)
fd.plot()
plt.show()
Importing data#
Usually the data used in the analysis comes from previous measurements of an experiment, that have been stored in some file format, such as comma-separated values (CSV), attribute-relation file format (ARFF) or Matlab and R formats. Users should know the particular layout used for representing functional data on their files, as there is currently no standard format for storing this kind of data.
Note
scikit-fda does not offer input/output functions right now. You can parse the grid points and values of the functions using the available tools in the Scientific Python ecosystem:
Numpy: for simple text-based or binary formats, such as CSV.
Scipy: for Matlab, Fortran or ARFF files.
RData: for loading data in the R file formats.
Pandas: includes tools for reading a variety of file formats, such as CSV, JSON, HTML, LaTeX, XML, Excel, HDF5, Parquet, SPSS or SQL.
Once the data is loaded as NumPy arrays, you can construct the
FDataGrid as explained above.
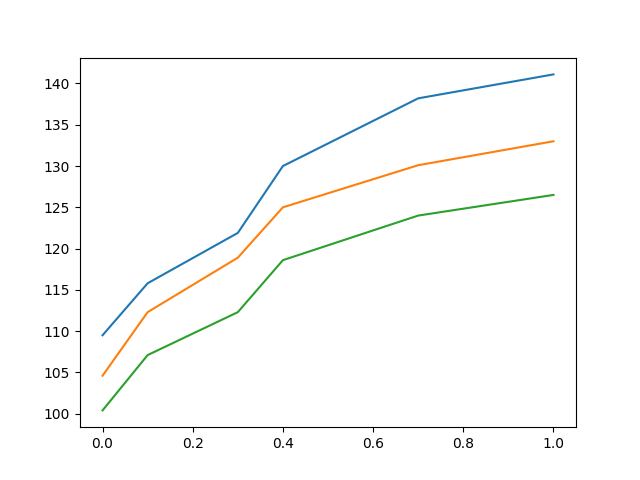
For example, consider the following file, containing unidimensional functional data in CSV form. The first row (the header) contains the grid points, common for all observations. Each of the following rows is a functional observation.
import pathlib
import textwrap
file_content = textwrap.dedent(
"""\
0.0, 0.1, 0.3, 0.4, 0.7, 1.0
109.5, 115.8, 121.9, 130.0, 138.2, 141.1
104.6, 112.3, 118.9, 125.0, 130.1, 133.0
100.4, 107.1, 112.3, 118.6, 124.0, 126.5
""",
)
test_file = pathlib.Path("data.csv")
test_file.write_text(file_content)
text = test_file.read_text()
print(text)
0.0, 0.1, 0.3, 0.4, 0.7, 1.0
109.5, 115.8, 121.9, 130.0, 138.2, 141.1
104.6, 112.3, 118.9, 125.0, 130.1, 133.0
100.4, 107.1, 112.3, 118.6, 124.0, 126.5
We can now load the CSV using the functions in Pandas.
Note that by default, Pandas reads the header as text and uses it to name
the columns.
Thus, we need to convert it to float before we can pass it to the
FDataGrid constructor.
import pandas
data = pandas.read_csv("data.csv")
grid_points = data.columns.astype(float)
data_matrix = data
We can now construct the FDataGrid
as before and plot it:
fd = skfda.FDataGrid(
data_matrix=data_matrix,
grid_points=grid_points,
)
fd.plot()
plt.show()
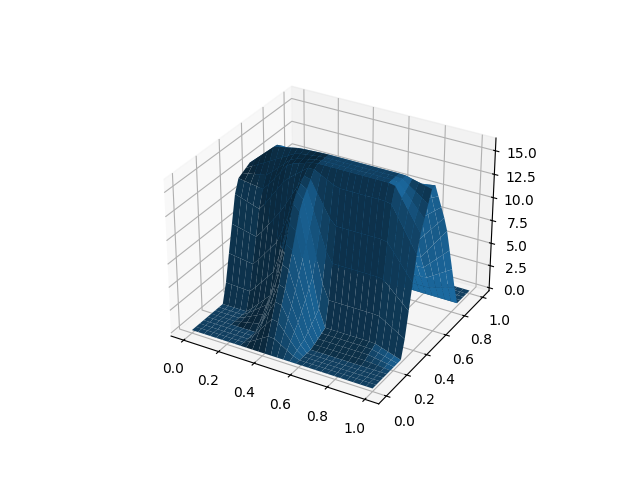
If you have data that is already a NumPy array, you can use it directly.
For example, we will load the
digits dataset of scikit-learn, which
is a preprocessed subset of the MNIST dataset, containing digit images.
The data is already a NumPy array. As the data has been flattened into a 1D
vector of pixels, we need to reshape the arrays to their original 8x8 shape.
Then this array can be used to construct the digits as surfaces.
from sklearn.datasets import load_digits
X, y = load_digits(return_X_y=True)
X = X.reshape(-1, 8, 8)
fd = skfda.FDataGrid(X)
# Plot the first 2 observations
fd[0].plot()
fd[1].plot()
plt.show()
Common datasets#
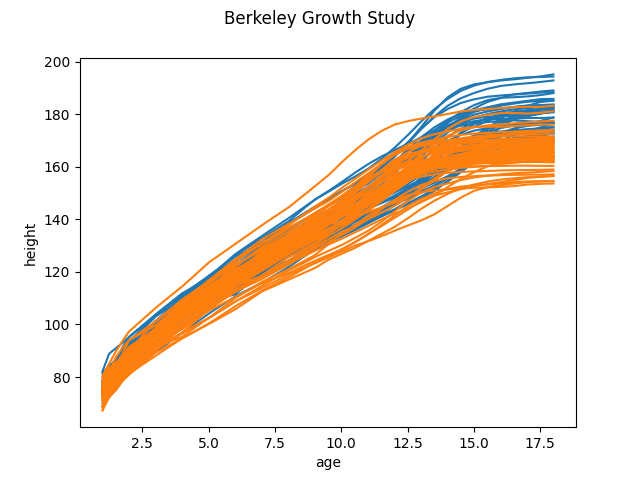
scikit-fda can download and import for you several of the most popular
datasets in the FDA literature, such as the Berkeley Growth
dataset (function fetch_growth()) or the Canadian
Weather dataset (function fetch_weather()). These
datasets are often useful as benchmarks, in order to compare results
between different algorithms, or simply as examples to use in teaching or
research.
X, y = skfda.datasets.fetch_growth(return_X_y=True)
X.plot(group=y)
plt.show()
Datasets from CRAN#
If you want to work with a dataset for which no fetching function exist, and
you know that is available inside a R package in the CRAN repository, you
can try using the function fetch_cran(). This function
will load the package, fetch the dataset and convert it to Python objects
using the packages
scikit-datasets and
RData. As datasets in CRAN follow no
particular structure, you will need to know how it is structured internally
in order to use it properly.
Note
Functional data objects from some packages, such as
fda.usc
are automatically recognized as such and converted to
FDataGrid instances. This
behaviour can be disabled or customized to work with more packages.
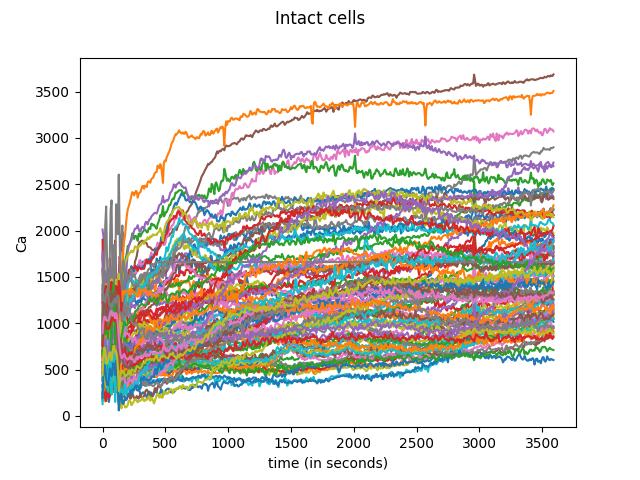
data = skfda.datasets.fetch_cran("MCO", "fda.usc")
data["MCO"]["intact"].plot()
plt.show()
Datasets from the UEA & UCR Time Series Classification Repository#
The UEA & UCR Time Series Classification Repository is a popular repository
for classification problems involving time series data. The datasets used
can be considered also as functional observations, where the functions
involved have domain dimension of 1, and the grid points are
equispaced. Thus, they have also been used in the FDA literature.
The original UCR datasets are univariate time series, while the new UEA
datasets incorporate also vector-valued data.
In scikit-fda, the function fetch_ucr() can be used
to obtain both kinds of datasets as
FDataGrid instances.
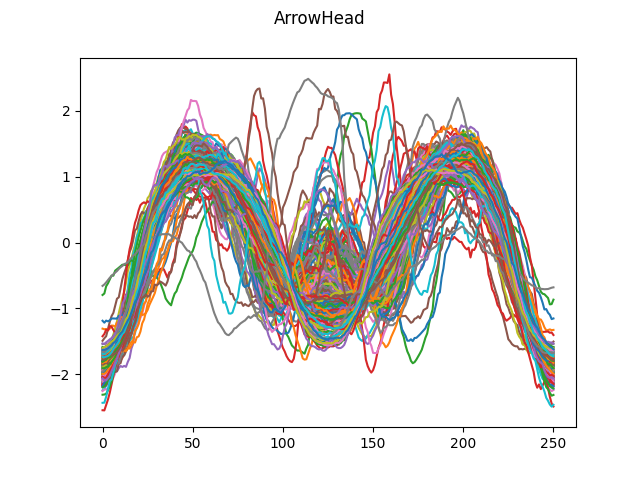
# Load ArrowHead dataset from UCR
dataset = skfda.datasets.fetch_ucr("ArrowHead")
dataset["data"].plot()
plt.show()
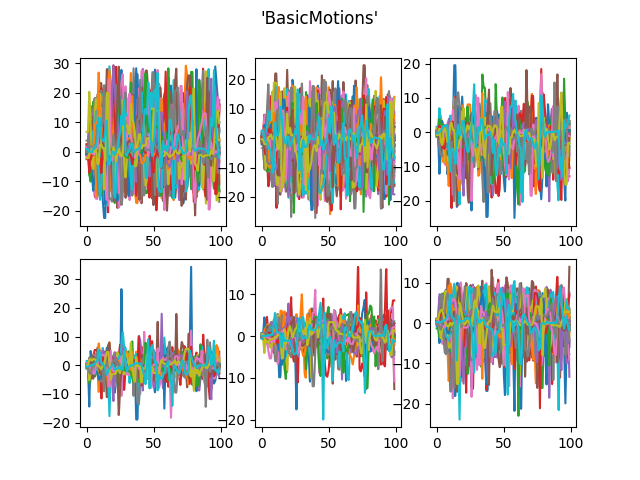
# Load BasicMotions dataset from UEA
dataset = skfda.datasets.fetch_ucr("BasicMotions")
dataset["data"].plot()
plt.show()
Synthetic data#
Sometimes it is not enough to have real-world data at your disposal. Perhaps the messy nature of real-world data makes difficult to detect when a particular algorithm has a strange behaviour. Perhaps you want to see how it performs under a simplified model. Maybe you want to see what happens when your data has particular characteristics, for which no dataset is available. Or maybe you only want to illustrate a concept without having to introduce a particular set of data.
In those cases, the ability to use generated data is desirable. To aid this
use case, scikit-learn provides several functions that generate data
according to some model. These functions are in the
datasets module and have the prefix make_.
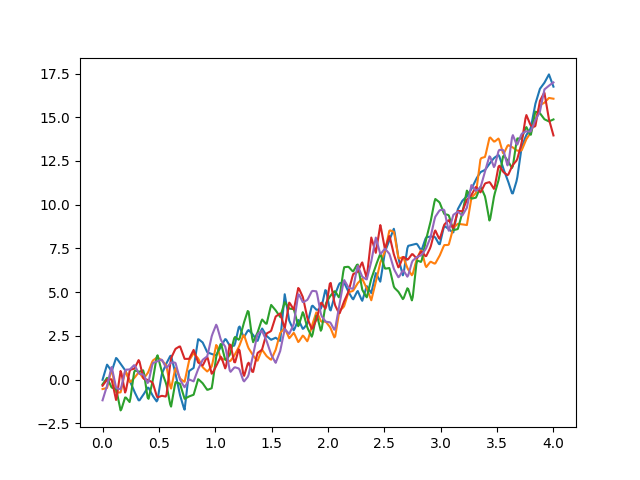
Maybe the most useful of those are the functions
skfda.datasets.make_gaussian_process() and
skfda.datasets.make_gaussian() which can be used to generate Gaussian
processes and Gaussian fields with different covariance functions.
cov = skfda.misc.covariances.Exponential(length_scale=0.1)
fd = skfda.datasets.make_gaussian_process(
start=0,
stop=4,
n_samples=5,
n_features=100,
mean=lambda t: t**2,
cov=cov,
)
fd.plot()
plt.show()
In order to know all the available functionalities to load existing and synthetic datasets it is recommended to look at the documentation of the datasets module.
Total running time of the script: (0 minutes 6.735 seconds)