Polynomial#
- class skfda.misc.covariances.Polynomial(*, variance=1, intercept=0, slope=1, degree=2)[source]#
Polynomial covariance function.
The covariance function is
\[K(x, x') = \sigma^2 (\alpha x^T x' + c)^d\]where \(\sigma^2\) is the scale of the variance, \(\alpha\) is the slope, \(d\) the degree of the polynomial and \(c\) is the intercept.
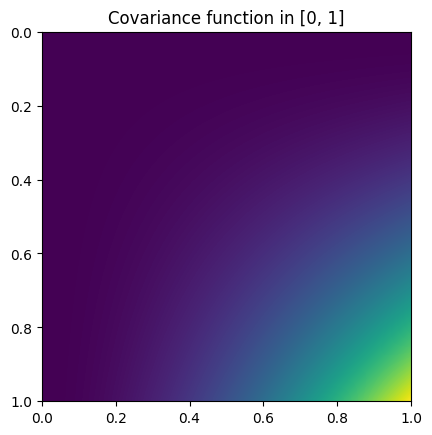
Heatmap plot of the covariance function:
from skfda.misc.covariances import Polynomial import matplotlib.pyplot as plt Polynomial().heatmap(limits=(0, 1)) plt.show()
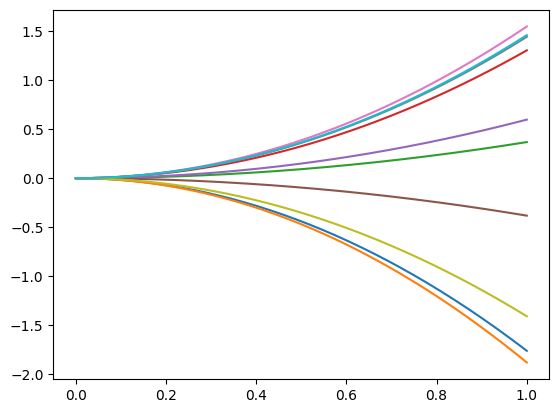
Example of Gaussian process trajectories using this covariance:
from skfda.misc.covariances import Polynomial from skfda.datasets import make_gaussian_process import matplotlib.pyplot as plt gp = make_gaussian_process( n_samples=10, cov=Polynomial(), random_state=0) gp.plot() plt.show()
Default representation in a Jupyter notebook:
from skfda.misc.covariances import Polynomial Polynomial()
\[K(x, x') = \sigma^2 (\alpha x^T x' + c)^d \\\text{where:}\begin{aligned}\qquad\sigma^2 &= 1 \\c &= 0 \\\alpha &= 1 \\d &= 2 \\\end{aligned}\]Methods
heatmap([limits])Return a heatmap plot of the covariance function.
Convert it to a sklearn kernel, if there is one.
